

Published on June 9, 2025, by Admin in the Luo Lab

In biological systems, cells precisely regulate gene expression. Individuals inherit genes from both parents, but alleles from different parents often exhibit differences in expression levels or patterns, which is known as Allele-Specific Expression (ASE). ASE may arise from two main causes: first, DNA sequence differences between alleles can affect their ability to bind regulatory elements; second, parent-of-origin effects, for which gene expression depends on whether the allele was inherited from the father or the mother. Genomic imprinting is the most typical example of a parent-of-origin effect, where genes are "imprinted" to express only the paternal or maternal allele. This imbalance in allele expression, caused by sequence differences or parent-of-origin effects (particularly genomic imprinting), is crucial for mammalian processes such as embryonic development, placental formation, growth metabolism, and neurological function. Disruption of this finely tuned regulation, such as abnormal expression of imprinted genes, often leads to severe developmental issues or even embryonic lethality.
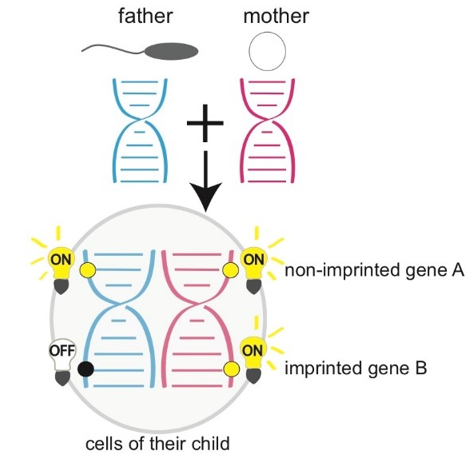
In addition to the traditional epigenetic mechanisms such as DNA methylation and histone modifications that regulate allele-specific gene expression, scientists are continuously exploring new regulatory layers. Among these, N6-methyladenosine (m6A)—the most prevalent post-transcriptional modification on mRNA and lncRNA in eukaryotes—has attracted significant attention in recent years. Much like a dynamic "chemical label" added to RNA molecules, m6A plays a crucial role in regulating various RNA processes, including splicing, nuclear export, translation, and degradation, thereby finely modulating gene expression.
Just as the expression of imprinted genes is strictly controlled by parent-of-origin effects, how are the expressions of alleles, which may have subtle sequence differences, precisely regulated across the genome? Does m6A, as a key RNA modification, also play a role in the regulation of allele-specific expression? Recently, our research team published a paper titled “Sequence and parent-of-origin dependent m6A contribute to allele-specific gene expression” in The EMBO Journal, focusing on these cutting-edge scientific questions. By sequencing the early brain tissues of two distantly related mouse strains (C57BL/6J and PWK/PhJ) and their F1 hybrid offspring, we systematically analyzed the distribution pattern of m6A modifications at single-base resolution on alleles, and their relationship with ASE.
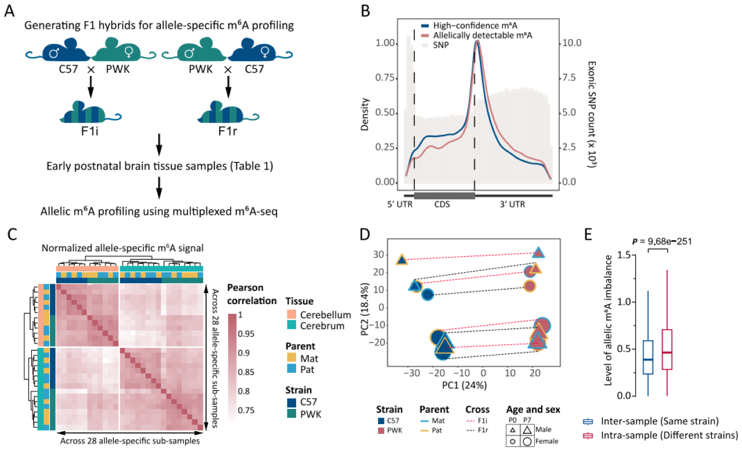
Based on our self-developed multi-sample parallelled m6A-RIP-seq, a quantitative detection technology, we identified thousands of m6A sites with significant modification level differences between alleles from different mouse strains. Further analysis revealed that these differences were primarily influenced by sequence variations near the alleles (such as SNPs). This suggests that the genetic sequence of the alleles plays a crucial role in determining their m6A modification patterns. Notably, the study uncovered a parent-of-origin effect for m6A modifications in known imprinted genes, which means that, even with identical sequences, alleles inherited from the father or mother may exhibit different m6A modification levels. Moreover, Whether sequence-dependent or parent-of-origin-dependent, the m6A allele-specific modifications observed in this study revealed a notable trend: alleles with higher m6A modification levels generally exhibit lower mRNA expression levels. This suggests that m6A modifications may negatively regulate the expression of specific alleles by modulating mRNA stability, such as by promoting degradation. This discovery further indicates that parent-of-origin information may not only regulate gene expression through traditional epigenetic mechanisms but also potentially through the regulation of RNA modifications, adding a new layer of complexity in the fine-tuned control of these developmentally critical genes. This expands the perspective on the regulatory mechanisms of genomic imprinting.

This study systematically unveils the crucial role of m6A RNA modifications in regulating allele-specific gene expression, offering a new molecular mechanism for understanding the complexity of gene expression regulation. Specifically, it establishes m6A modification as an important post-transcriptional regulator of allele expression differences, expanding our understanding of the ASE regulatory network. The negative regulation pattern of allele expression mediated by m6A offers new insights into gene dosage compensation, fine-tuned regulation during development, and the genetic basis of complex traits and diseases. These findings also provide new directions and potential intervention targets for the study of developmental disorders and disease mechanisms. Ying Zhang, Ze-Yu Zhang (from the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) and Hong-Xuan Chen are co-first authors of this paper. Professor Guan-Zheng Luo is the corresponding author. Luo Lab members, including Biao-Di Liu and Ye-Lin Lan, also contributed to the study. Congratulations to them! Additionally, this work received strong support from Professor Xiu-Jie Wang of the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences; Dr. Wei Li and Dr. Guihai Feng from the Institute of Zoology, Chinese Academy of Sciences.
By Ying Zhang and Hong-Xuan Chen