

Published on June 3, 2025, by Admin in the Luo Lab

On June 2nd, 2025, our lab achieved a significant breakthrough in single-molecule sequencing and RNA epitranscriptomics with our findings, titled “Single-molecule direct RNA sequencing reveals the shaping of epitranscriptome across multiple species”, published in Nature Communications. In this study, we developed a high-precision single-molecule m6A detection tool, SingleMod, based on nanopore direct RNA sequencing (DRS), and systematically portrayed the m6A single-molecule epitranscriptome across multiple human cell lines and distant species unprecedentedly. This work unveils a series of previously hidden m6A features and evolutionary patterns.
RNA methylation, particularly m6A modification, is a crucial regulatory mechanism of gene expression and plays a pivotal role in cellular differentiation, development, and disease processes. However, current m6A detection methods primarily rely on next-generation sequencing (NGS), which requires RNA fragmentation and reverse transcription. These methods provide mixed signals from multiple molecules, making it difficult to reconstruct the true modification status of each individual RNA molecule. Such "averaged" detection masks the inherent heterogeneity among RNA molecules and limits our understanding of the complexity of the epitranscriptome. Nanopore direct RNA sequencing offers a revolutionary opportunity for studying the epitranscriptome at the single-molecule level, yet previous computational tools have been unable to effectively detect m6A at this level (as detailed in our previous publication in Nature Communications, 2023). In this study, we proposed the Multi-instance Regression (MIR) algorithm, which efficiently utilizes the methylation rate labels from NGS-based absolute quantification methods for training, and developed SingleMod, a tool designed to predict m6A at the single-molecule level from DRS data. Through comprehensive validation using multiple orthogonal datasets, we demonstrated the outstanding performance and robustness of SingleMod in single-molecule m6A detection. Furthermore, SingleMod is compatible with both RNA002 and RNA004 DRS datasets for m6A detection.
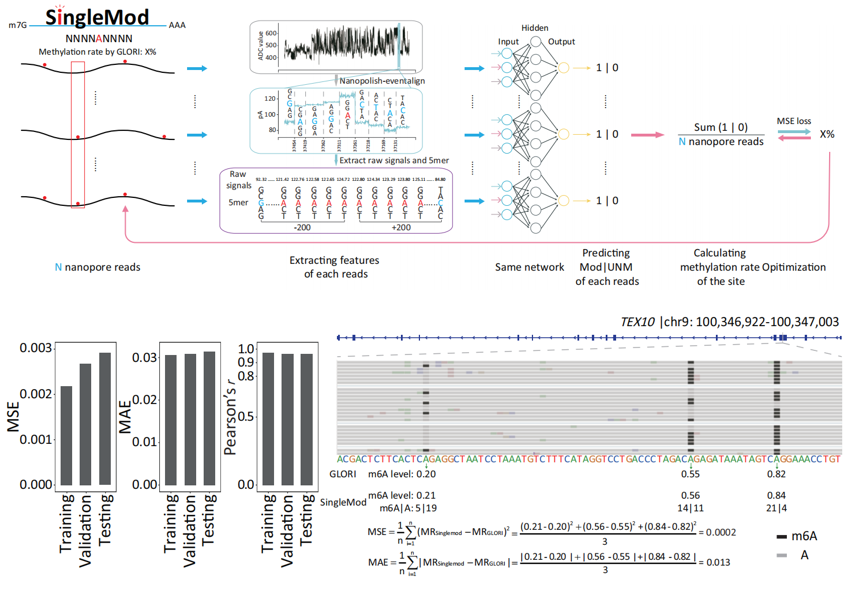
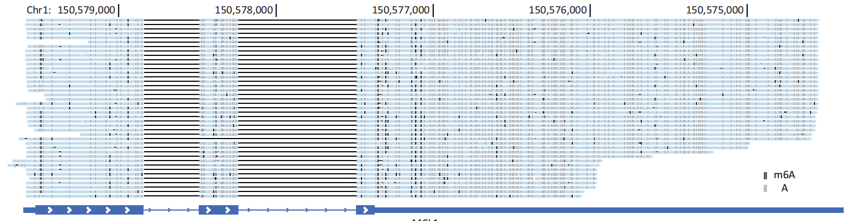
"Seeing" m6A Modifications on Each RNA Molecule——We applied SingleMod to depict m6A single-molecule epitranscriptomes in 3 human cell lines and, for the first time, investigated the fine quantification, intercellular stability, distribution characteristics, and molecular heterogeneity of m6A at the single-molecule level. Notably, we found that m6A significantly increased the heterogeneity of RNA molecules within the same gene or transcript.
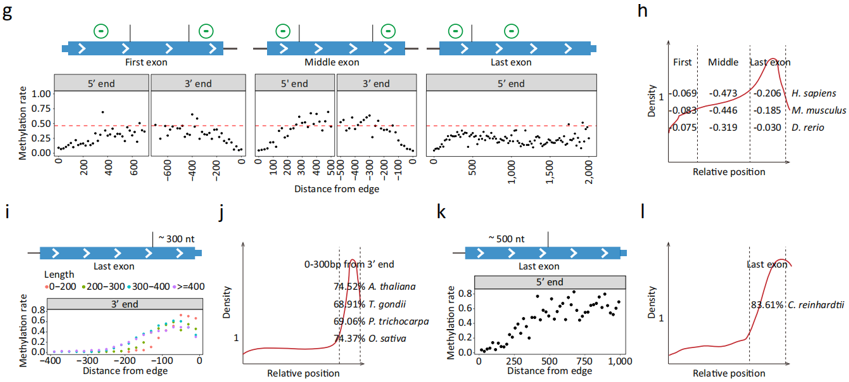
Conservation and Diversity of m6A Distribution——By systematically comparing the single-molecule m6A epitranscriptomes of 8 distant species, including mammals, fish, plants, protists, and green algae, we identified 3 distinct m6A distribution patterns: the vertebrate pattern, the plant and protist pattern, and the green algae pattern. Additionally, we discovered that the EJC-mediated exclusion model of m6A addition could not explain the m6A distribution patterns in non-vertebrate species, suggesting the existence of unique m6A regulatory mechanisms in these organisms.
In summary, this study not only achieves a key breakthrough in m6A modification detection technology but also provides a series of novel biological insights into m6A at the single-molecule level, offering a fresh perspective for epitranscriptome research. Ying-Yuan Xie, Zhen-Dong Zhong, and Hong-Xuan Chen are the co-first authors of this article, with Professor Guan-Zheng Luo and Associate Professor Zhang Zhang as the corresponding authors. Other members of Luo Lab, including Yuan-Tao Qiu and Ze-Hui Ren, also contributed to this work. Congratulations to them! Additionally, this study received significant support from Professor Jian Wu at South China Agricultural University, as well as Dr. Zi-Gang Li and Dr. Feng Yin at Shenzhen Bay Laboratory.
By Ying-Yuan Xie and Hong-Xuan Chen