

Published on May 30, 2023, by Admin in the Luo Lab

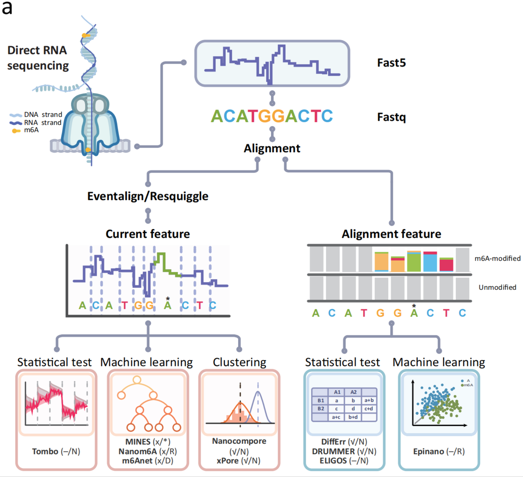
N6-methyladenosine (m6A) is the most abundant and thoroughly studied RNA modification base in vertebrates. Currently, m6A detection is mainly based on second-generation sequencing methods, which have problems such as false positives, complex operations, and limited to site detection. Gradually, nanopore sequencing technology (Oxford Nanopore Technologies, ONT) has become an ideal alternative method. Although many highly complex and advanced computational tools have been developed to detect and quantify m6A via nanopore sequencing, there is still a lack of research to thoroughly evaluate and compare these tools.
Our group published a paper entitled “Systematic comparison of tools used for m6A mapping from nanopore direct RNA sequencing” in Nature Communications on April 5, 2023. In this research,we used multi-species multi-replicate samples as evaluation data and comprehensively evaluated these tools for detecting m6A. Our results showed that there was a trade-off between accuracy and recall rate for most tools. In addition, we also evaluated the inherent bias of these tools during detection and found that introducing negative control samples can improve their performance. Detection ability varies between different motifs and sequencing depth and stoichiometry of modification are two other important factors. All in all, we provides in-depth insights into current computational tools used to detect m6A based on ONT DRS data and highlights the potential for further improvement of these tools.
Laboratory members Zhendong Zhong and Yingyuan Xie are co-first authors of the article. Hongxuan Chen, Yelin Lan, Xuehong Liu, Jingyun Ji and Fu Wu also contributed to this article. At the same time we received help from Prof. Jiekai Chen from Guangzhou Health Institute and Prof. Daniel W. Mak from Hong Kong University. Cheers!
By Guorun Tang